In an attempt to apply the idea of Reproducible Research to my work, I devised a Makefile-based project to reproduce data published by the NIST on a simple Lennard-Jones system.
Reproducible Research
Reproducible research consists in documenting a workflow (from parameters to figure generation) for the computer, so that a result can be reproduced. For a idea, here are a few interesting links: initial steps toward reproducible research, Baby steps for the Open-Curious or Ten Simple Rules for Reproducible Computational Research.
My goal here was to reproduce reference data published by the National Institute of Standards and Technology (NIST) as part of their benchmark simulations of Lennard-Jones fluids, in the "MD NVE" category. This seemed an appropriate project to test reproducible research as it is one of the most common and simple model for the Molecular Dynamics of fluids.
Run it
If you are interested in reproducing the following figure:
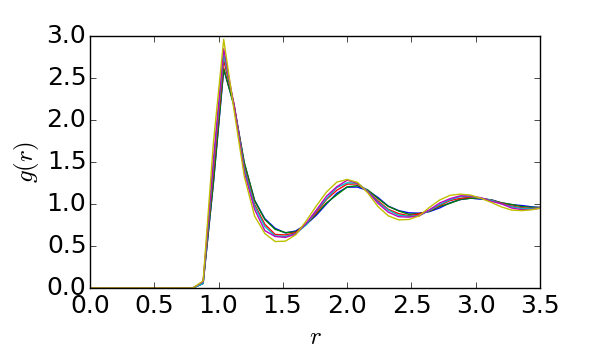
Install a bunch of software:
and a program called sftmpl (for "Single file templater") that is available
via pip
pip install sftmpl
or at https://github.com/pdebuyl/sftmpl. If you do not have installation
rights on your machine, you may use pip install --user sftmpl.
The custom dump style for H5MD is available at https://github.com/pdebuyl/lammps and is needed to take advantage of the analysis tools in ljrr.
Once the software is installed, the project is obtained via git
git clone https://github.com/pdebuyl/ljrr
cd ljrr
To reproduce the figure, invoke the make command.
make data/nist_rdf.png
Your computer should now stay busy for some time. It is running the MD simulations at the different parameter values of the benchmark simulations. For information, running on a single CPU (Intel Core i5 at 3.4 GHz), the whole process take about 24 minutes.
The process
What happens during this time?
- Make realizes that to build the figure it needs a program,
code/plot_rdf.pyand a series of datafiles. - The datafiles are computed by the program
code/compute_rdf.pybut they depend on the raw simulation data. - To obtain the raw simulation data, make invokes
lmp_mpi(the lammps executable). The configuration file for the simulations is generated by the programsftmplfrom the templatein.lj3d.tmplby filling the appropriate variables with parameters.
The execution of lammps is repeated by a bash program, run_until_lj3d.sh until
the check_T.py program confirms that the temperature is whithin a small range
around 0.85. The bash program also generates a new seed at each time from the
/dev/urandom device of your computer.
Additional remarks
Here are a few remarks: the principles I followed and some of the things I have learnt.
-
I don't know if the makefile is the most {simple,elegant,clean}. What I learnt is that loops and passing many parameters is not always practical with makefiles. What couldn't be done with the makefile was done with bash.
-
I could have automated the process with bash only (no dependency management then) or relied on Python but I wanted to remain generic with respect to the software actually doing the computations.
-
This project requires a lot of software (10 explicit dependencies). I suppose that it is typical of "real life" research projects in that sense. I must say that I use all of these components anyway but didn't realize that even what I use for a small project may not be installed on everyone's computer.
-
This project is about the production of data. In a future blog post I will explore the publication of said data using ActivePapers
-
I could have used lammps' feature to compute the radial distribution function. But again, I wanted to be generic so that the process could be applied to the output of any simulation code.
Comments welcome via twitter or by email (pdebuyl at domainname of this blog).
Comments !
Comments are temporarily disabled.